Data Models¶
PyUniProt uses SQLAlchemy to store the data in the database.
Use instance of pyuniprot.manager.query.QueryManager to query the content of the database.
Entity–relationship model:
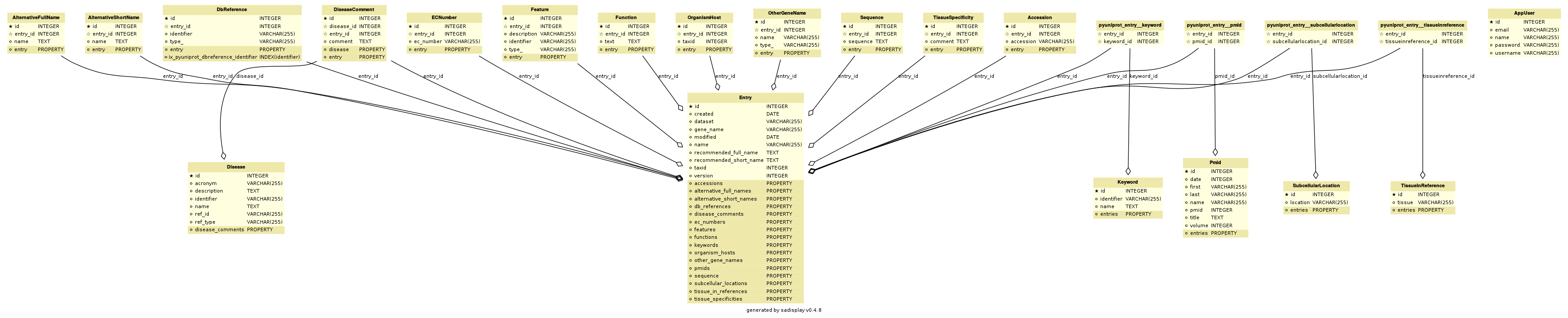
Contents
Entry¶
-
class
pyuniprot.manager.models.Entry(**kwargs)[source]¶ UniProt entry with relations to other models
Query
Entrywithpyuniprot.manager.query.QueryManager.entry()Variables: - dataset (str) – dataset type (SwissProt or TrEMBL)
- created (datetime.datetime) – Date created
- modified (datetime.datetime) – Datemodified
- version (int) – Dataset version
- name (str) – UniProt entry name
- recommended_full_name (str) – Recommended full protein name
- recommended_short_name (str) – Recommended short protein name
- taxid (int) – NCBI taxonomy identifier
- gene_name (str) – Primary gene name
- sequence – 1:1 to
Sequence - accessions (collections.Iterable) – 1:n to
Accession - organism_hosts (collections.Iterable) – 1:n to
OrganismHost - features (collections.Iterable) – 1:n to
Feature - functions (collections.Iterable) – 1:n to
Function - ec_numbers (collections.Iterable) – 1:n to
ECNumber - db_references (collections.Iterable) – 1:n to
DbReference - alternative_full_names (collections.Iterable) – 1:n to
AlternativeFullName - alternative_short_names (collections.Iterable) – 1:n to
AlternativeShortName - disease_comments (collections.Iterable) – 1:n to
DiseaseComment - tissue_specificities (collections.Iterable) – 1:n to
TissueSpecificity - other_gene_names (collections.Iterable) – 1:n to
OtherGeneName - pmids (collections.Iterable) – n:m to
Pmid - keywords (collections.Iterable) – n:m to
Keyword - subcellular_locations (collections.Iterable) – n:m to
SubcellularLocation - tissue_in_references (collections.Iterable) – n:m to
TissueInReference
Table view
Links
For more information on UniProt website:
OtherGeneName¶
Pmid¶
Feature¶
-
class
pyuniprot.manager.models.Feature(**kwargs)[source]¶ Sequence annotations describe regions or sites of interest in the protein sequence, such as post-translational modifications, binding sites, enzyme active sites, local secondary structure or other characteristics reported in the cited references. In the moment we don’t save the positions. If this is strongly neede contact the PyUniProt team on github.
Variables: Table view

Links













